![]() Classification and free energy landscape of entanglement proteins: knots, slipknots and lassos
Classification and free energy landscape of entanglement proteins: knots, slipknots and lassos
Białka splątane – studium nowych struktur i rozwiązanie ich zagadki
Ideas Plus II grant, MNiSW
This project is devoted to a fascinating subject of entanglement in proteins. Within the last few years it has been realized that proteins may form non-trivial entangled structures, such as knots, slipknots or tadpoles, whose existence is obscured in a standard representation in terms primary, secondary, tertiary (and other) structures. While analysis of knotted proteins has already grown into an independent and rapidly developing field, with this project we are going to go beyond its borders and include other tangled structures – complex lasso proteins and links, into analysis. The main goal of this project is to understand energy landscape of such tangled structures based on mechanical manipulations (Hummer, G et al.), (Sulkowska JI, et al.), (Sulkowska JI, et al.) and their function, by means of theoretical models, whose predictions will be supported and verified in experiment.
![]() Influence of knotted structure on function of proteins and protein structure prediction
Influence of knotted structure on function of proteins and protein structure prediction
Sonata BIS grant, NCN
The main goal of this project is to understand the role and function of knots in proteins. Our research hypothesis assumes that existence of knots (Mansfield ML, “Are there knots in proteins?” (1994) Nature Struct Biol 1(4):213‐214), and other nontrivial geometric structures such as so-called slipknots (Sulkowska JI, et al.), is correlated with functional properties of proteins (Sulkowska JI, et al.). In particular, we expect that geometric structure of knots to be correlated with the location of active sites in proteins; moreover knotted structures should be manifested in molecular motion of proteins, as well as in existence of co-evolving pairs of amino-acids. As knotted proteins are extremely difficult to study experimentally, it is crucial to develop first theoretical approach to study their function – and this is the main approach that we are going to focus on.
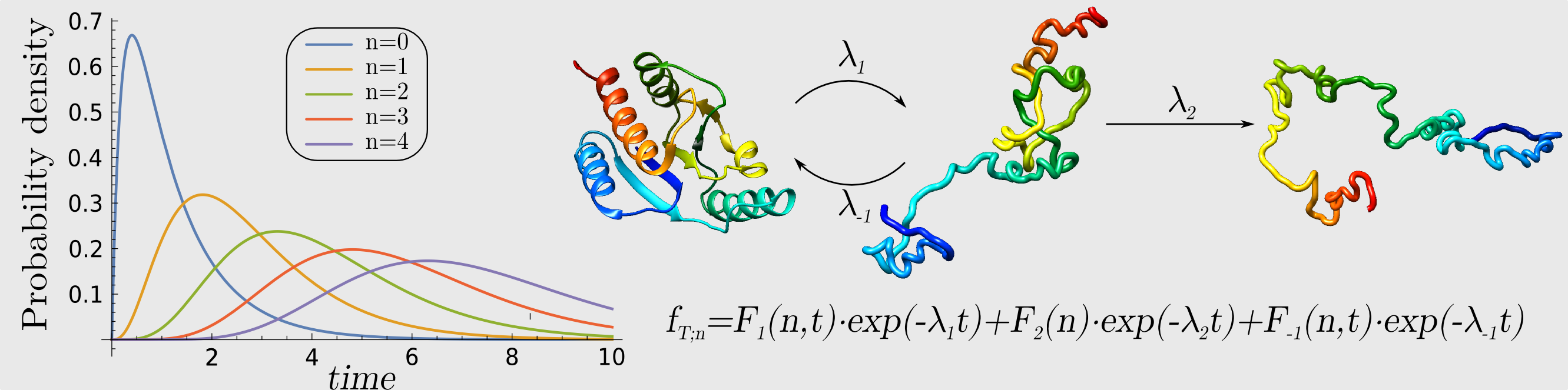
Comprehensive analysis of knotted proteins 
EMBO Installation Grant
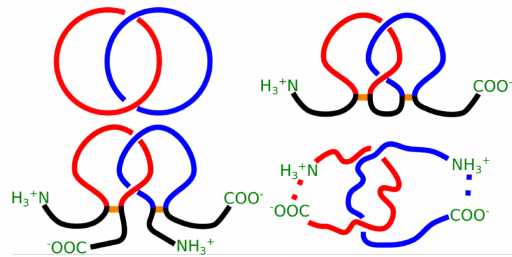
To perform some function we can combine many independent parts together and create a machine with all those parts incorporated as in a Swiss watch. Alternatively, we can consider a single, albeit more complicated object, which can perform the same function. A remarkable new paradigm emerging from recent biophysical studies is related to this latter mechanism, which appears to be used in Nature to perform several complicated biological reactions. In this case, the role of a single, more complicated object is played by a non-trivial topology of a protein. By “non-trivial topology”, as it is used in a mathematical sense, we mean here a knotted configuration of a protein backbone – i.e. pulling such a protein by its termini would result in a tightened knot (Sulkowska JI, et al. PNAS 2012). A related interesting structure is so called the “slipknot” (King NP, et al., Sulkowska JI, et al.). In contrast, a “trivial topology” denotes a protein configuration without a knot, whose pulling would result in an unfolded chain (Sulkowska JI, et al.) For example, a process of methylation in archaea (tRNA ribose 2′-O-methylase) is performed by complex dependent on other protein interactions and guidance from RNA (C/D sRNP), whereas in eukaryotes it is performed just by one enzyme (Lahoud G, et al.). Apparently, it has been found that this enzyme is knotted (Elkins PA, et al.). The role of a knot in this enzyme clearly deserves explanation. The main aim of this project is to understand, by means of computer simulations and experimental techniques, the role played by knots in proteins in various processes. Our achievement in this topic is summarized here (Christian T, Sakaguchi R, Perlinska AP, et al.).
 Can biology be caught on a mathematical lasso?
Can biology be caught on a mathematical lasso?
Czy biologię da się schwytać na matematyczne lasso?
Inter, FNP – popular science grant – Wanda Niemyska
The aim of this project is to use the theory on the borderline of mathematics and physics regarding certain characteristics of curves and surfaces in three-dimensional space to create tools to uncover the process of formation (folding) of recently discovered structures with new type of topologies in proteins, resembling the shape of a lasso (Niemyska, W et al.).
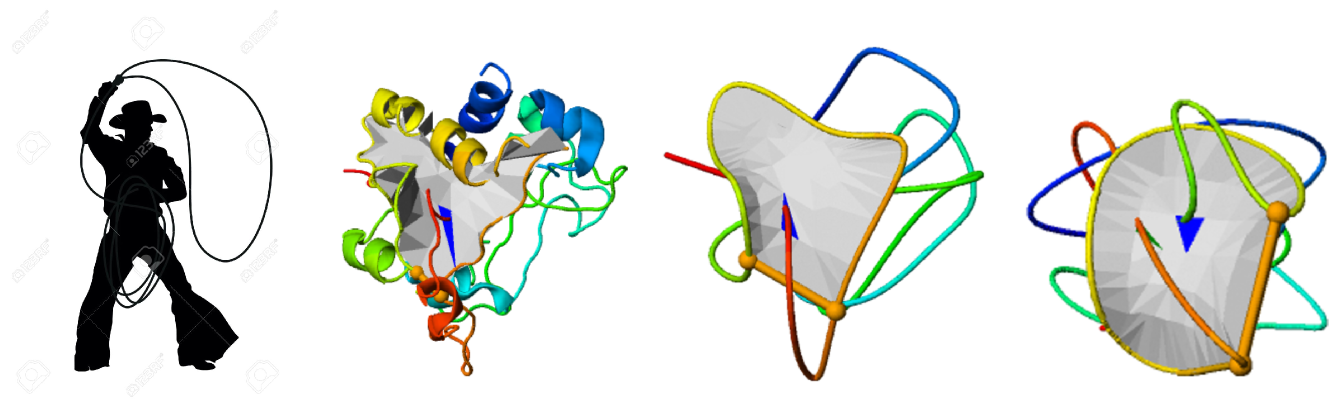
 Characterization of the structure-function relation in proteins with lassos
Characterization of the structure-function relation in proteins with lassos
Charakteryzacja zależności struktura-funkcja w białkach o topologii typu lasso
Grant for young researchers (DSM) – Paweł Dąbrowski-Tumański
Synthesis of inhibitors for TrmD proteins” 
EMBO Small Grant
Our goal is to find selective inhibitor for a knotted TrmD tRNA methyltransferase. The first step is to synthesize and test three ligands suggested previously as selective by Astra Zeneca company. Project in collaboration with Jacek Jemielity group.
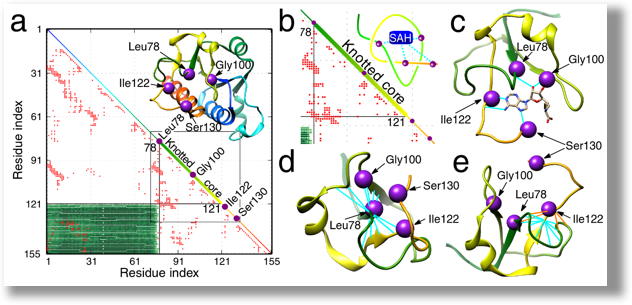
Great challenges
ICM computational grant
The main goal of the project is to investigate the influence of non-trivial topology on ligand binding based on TrmD family, in order to understand the role of entanglement in the biological function of proteins. Investigated proteins are characterized by a knot they form. Such a knot forms an active site, and thus it can directly affect the function of a protein through interaction with ligands. These studies are carried out in close cooperation with a group of experimenters (group of prof. Ya-Ming Hou, Thomas Jefferson University, USA), who determined structures of protein with ligands and tRNA (Ito, Takuhiro, et al.)
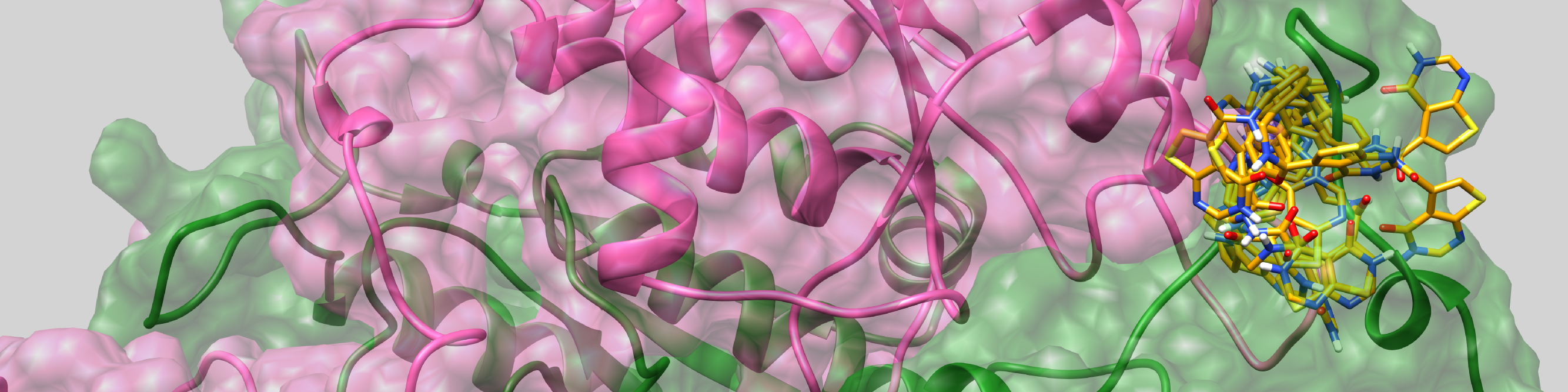
 Development of a methodology to uncover the macrocyclic links in proteins
Development of a methodology to uncover the macrocyclic links in proteins
Opracowanie metodologii wykrywania splotów makrocykliczych w białkach
Grant for young researchers (DSM) – Paweł Dąbrowski-Tumański
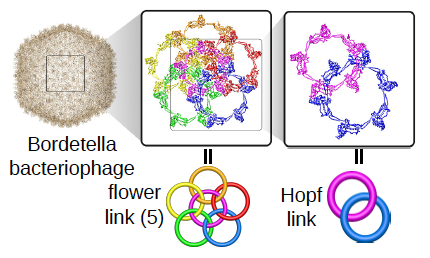
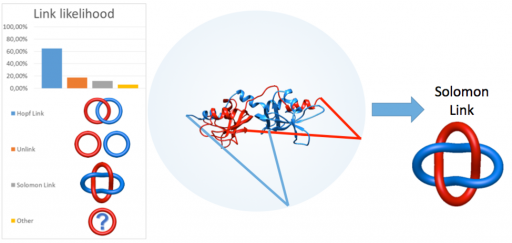
![]() Identification and characterization of proteins with link topology
Identification and characterization of proteins with link topology
Identyfikacja i charakteryzacja białek o topologii splotu
Preludium grant, NCN – Paweł Dąbrowski-Tumański
The aims of the project are development of new methods aiming to automatically detect various types of link topology in protein structure, topological classification and creation of publicly available server and database (the LinkProt database), analysis of folding and determination of the free energy landscape of link proteins and search for the influence of the topology on function and properties of proteins. To study folding of linked proteins the appropriate model enabling to conduct fast and reliable simulations of protein dynamics in different conditions will be developed. The topological classification will involve utilization of mathematical tools from knot theory and theory of minimal surfaces.