Stay Tuned for Upcoming Opportunities!
We regularly have new positions opening. Please check back soon, visit the lab’s Facebook page or contact us! Below some previous opportunities are listed.
2 POSTDOCS
Application deadline: 31 August 2019
Position duration: 1 to 3 years, from 2 September 2019
Salary: 8 000 – 10 000 PLN monthly
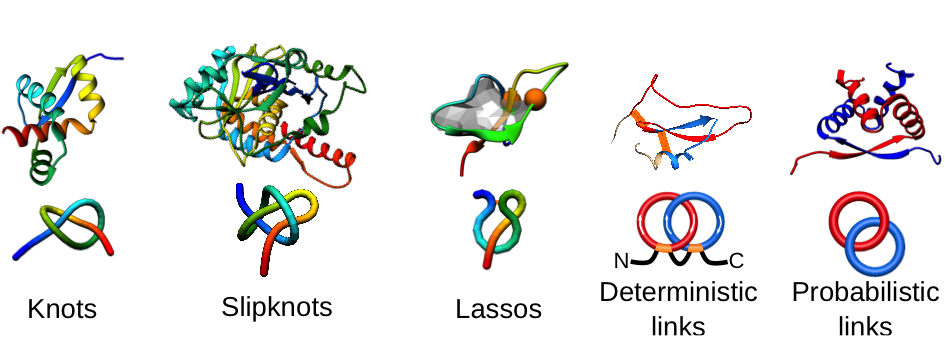 The goal of this project is to develop new methods to apply experimental data from NMR and optical traps to expand our knowledge of proteins with non-trivial topology. Currently, it is still not known how to untie experimentally knotted proteins by chemical or thermal denaturation. Therefore our goal is to determine their function and the folding process (the free energy landscape) through the single-molecule level approach (AFM, optical traps). Moreover, to fully understand these exotic structures, their degradation in proteasome will be investigated via the NMR (in deuterated water). The role of the applicant will be to include these data into computational study. Thus, through the use of multiscale computational methods, we will investigate on one hand the conformational stability, the strength of the internal interactions, as well as routes to tie/untie proteins, and on the other hand, we will use state-of-the-art methods to take into account non-trivial topology of investigated proteins. In this work, we will use a combined coarse grained and explicit solvent molecular dynamics / replica exchange approaches and statistical mechanics methods to determine the unbiased landscape of proteins with non-trivial topology.
The goal of this project is to develop new methods to apply experimental data from NMR and optical traps to expand our knowledge of proteins with non-trivial topology. Currently, it is still not known how to untie experimentally knotted proteins by chemical or thermal denaturation. Therefore our goal is to determine their function and the folding process (the free energy landscape) through the single-molecule level approach (AFM, optical traps). Moreover, to fully understand these exotic structures, their degradation in proteasome will be investigated via the NMR (in deuterated water). The role of the applicant will be to include these data into computational study. Thus, through the use of multiscale computational methods, we will investigate on one hand the conformational stability, the strength of the internal interactions, as well as routes to tie/untie proteins, and on the other hand, we will use state-of-the-art methods to take into account non-trivial topology of investigated proteins. In this work, we will use a combined coarse grained and explicit solvent molecular dynamics / replica exchange approaches and statistical mechanics methods to determine the unbiased landscape of proteins with non-trivial topology.
For details on requirements, tasks and how to apply please consult the PDF file.
2 STUDENTS
Position duration: up to 24 months, from 1 October 2019 or earlier
Studentship: 1 000 – 1 500 PLN monthly
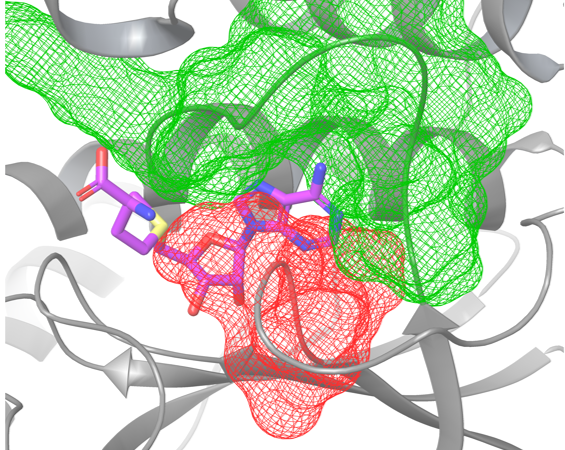 Celem projektu jest zaprojektowanie nowych antybiotyków mających działać poprzez inhibicję TrmD. TrmD to metylotransferaza tRNA, będąca jednym z kluczowych enzymów dla bakterii takich jak np. Haemophilus influenzae czy Streptococcus pneumoniae. Jedną z głównych trudności przy projektowaniu nowych inhibitorów TrmD jest występowanie spokrewnionego enzymu Trm5 w ludzkim organizmie. Utrudnia to uzyskanie selektywności potencjalnych ligandów, a przez to stwarza ryzyko toksyczności. Kluczową różnicą między tymi enzymami jest występowanie węzła w miejscu aktywnym TrmD, w przeciwieństwie do Trm5. Daje to możliwość projektowania selektywnych inhibitorów enzymu bakteryjnego.
Celem projektu jest zaprojektowanie nowych antybiotyków mających działać poprzez inhibicję TrmD. TrmD to metylotransferaza tRNA, będąca jednym z kluczowych enzymów dla bakterii takich jak np. Haemophilus influenzae czy Streptococcus pneumoniae. Jedną z głównych trudności przy projektowaniu nowych inhibitorów TrmD jest występowanie spokrewnionego enzymu Trm5 w ludzkim organizmie. Utrudnia to uzyskanie selektywności potencjalnych ligandów, a przez to stwarza ryzyko toksyczności. Kluczową różnicą między tymi enzymami jest występowanie węzła w miejscu aktywnym TrmD, w przeciwieństwie do Trm5. Daje to możliwość projektowania selektywnych inhibitorów enzymu bakteryjnego.
For details on requirements, tasks and how to apply please consult the PDF file.